Abstract
Introduction:
Despite major treatment improvements over the past decades, pediatric acute myeloid leukemia (AML) is still a life-threatening malignancy with relapse rates up to 30% and survival rates below 75%. Moreover, most of investigations are based on the study of large cohorts of adult AML patients while genetic profiles are known to be quite different between adults and children with AML. In this context, a better description of the pattern of molecular aberrations in childhood AML remains a great challenge to refine prognostication and improve outcome in such patients. We report here the comprehensive molecular landscape of a large and well-annotated cohort of pediatric AML and propose a new prognostic molecular classifier in this particular group of patients.
Patients and methods:
We performed molecular analysis using both high-throughput sequencing focused on 36 genes recurrently mutated in myeloid malignancies and ligation-dependent RT-PCR detecting more than 50 recurrent gene rearrangements in 385 patients with AML of the 438 children enrolled in the prospective ELAM02 trial and we evaluated their prognostic significance.
Results:
The median age at AML diagnosis was 8.6 years (range: 0-18) and the median WBC count was 16.6×109/L (range: 0.40-575). The distribution in the cytogenetic subgroups was as follows: normal karyotype (n=101, 26.2%), CBF-rearranged (n=92, 24% including t(8;21): n=57 and inv(16)/t(16;16): n=35), KMT2A (MLL)-rearranged (n=79, 21%), adverse karyotype (n=40, 10% including complex karyotype: n=27, monosomy 7: n=9 and t(6;9): n=4) and other (n=73, 19%). 76% of patients had at least one driver mutation among the genes we screened. The most common class of mutations involved genes that control tyrosine kinase signaling (61% including CBL, FLT3, JAK2, KIT, KRAS, MPL, NRAS, PTPN11, SETPB1) followed by transcription factors (16%: CEBPA, ETV6, GATA1, GATA2, RUNX1), tumor suppressors (14%: PHF6, PTEN, TP53, WT1), NPM1 (9%), chromatin modifiers (9%: ASXL1, BCOR, BCORL1, EZH2), DNA methylation controllers (8%: DNMT3A, IDH1, IDH2, TET2), cohesin genes (5%: NIPBL, RAD21, SMC1A, SMC3, STAG2) and spliceosome (3%: SF3B1, SRSF2, U2AF1, ZRSR2). Moreover, a recurrent transcript fusion was detected in about a half of pediatric patients. Taken together, we identified at least one molecular aberration (mutations or fusion transcripts) in 344 (89%) out of 385 patients.
In multivariate analysis, the presence of a NUP98 fusion is the sole variable associated with induction failure (50% CR, P=0.038). NUP98 -rearranged cases represented 2.6% of this cohort (10 patients/385 in which 5 had a normal karyotype). When studying OS, 5 factors revealed a negative impact : (1) WBC count higher than 30×109/L (P=0.001); (2) WT1 mutations (P=0.027); (3) RUNX1 mutations (P=0.043); (4) PHF6 mutations (P=0.038) and (5) adverse cytogenetics (P<0.001). On the other hand, 3 factors were shown to positively impact OS: (1) NPM1 mutations (P<0.001), (2) CEBPA dm (P=0.042) and (3) CBF-rearrangements (P<0.001).
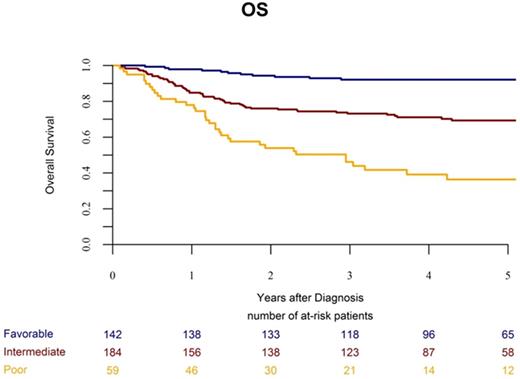
Considering results from multivariate analysis and strong molecular markers validated among studies, we defined a molecular classifier, refining the prognosis in childhood AML (see figure). Overall, CBF rearrangements, NPM1 and double CEBPA mutations represented 37% of the cohort and defined a favorable molecular subgroup (3-years OS: 92%) while NUP98 fusions, WT1, RUNX1 and PHF6 mutations (15% of the cohort) segregated into a poor molecular subgroup (3-years OS: 46%). Neither karyotype nor others gene mutations were able to discriminate patients into the intermediate molecular risk subgroup (3-years OS: 73%). KMT2A -rearrangements (21% of the cohort) were associated with an intermediate risk.
Conclusion:
Despite some overlaps, the spectrum of molecular aberrations and their prognostic significance differ between childhood and adult AML. These results have important implications to contribute in refining risk stratification of pediatric AML and show the need for further validations in independent pediatric cohorts.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal